DNA Isolation
- Before any type of testing can be performed DNA must be separated from the rest of the cellular components, as well as from any non-biological material that may be present.
- Foreign material may affect efficiency of DNA enzymes used in procedures.
- Extraneous substances can cause DNA to degrade
- DNA isolation procedure varies according to:
- Type of biological evidence present (blood, semen, salvia, hair).
- Amount of evidence.
- Kinds of cells present.
- Chelex Extraction
- Used on minute samples eg. Speck of barely visible blood.
- Sample boiled in a solution containing minute beads of a chemical called chelex.
- Boiling breaks open the cells, releasing the DNA and Chelex binds up most of the other extraneous materials.
- Chelex beads are removed along with most non-DNA components, leaving behind the DNA.
- This technique breaks apart double stranded DNA so is not suitable for RFLP analysis where dsDNA is needed, but PCR analysis can be performed on ssDNA.
- This system not suitable for STR analysis.
- QiaAmp Extraction
- This is a silica column-based method and is used to remove DNA from other cellular components after cells are lysed.
- The DNA extract is passed over a small column containing miniscule beads composed of a substance to which DNA adheres under certain chemical conditions. All other material is washed off the column. The chemical conditions are then changed to elute the DNA.
- DNA isolated by this method is double stranded and of high quality.
- Organic Extraction
- In this procedure the sample is cut into small pieces and soaked in a warm solution to gently release the cells from the substrate on which they are deposited.
- Another chemical plus mild heat is then used to break open cells, releasing DNA.
- The DNA is then isolated using various organic solvents.
- DNA further purified and concentrated.
- Differential Extraction
- Procedure used to isolate DNA from a mixed sample of sperm and non-sperm cells.
- The different properties of sperm from other cells (Including: saliva, skin, buccal and vaginal cells), is exploited to separate them from others before DNA extraction.
- All cells are first removed from the substrate by soaking in a gentle solution.
Determining Quantity and Quality of DNA
- Quantity
- Forensic samplesfrequently contain DNA co-extracted from microbial organisms such as yeasts and bacteria.
- If RFLP analysis is used it is necessary to consider both total DNA and proportion of human DNA.
- The proportion of human DNA determines how much sample to load on the analytical gel.
- For RFLP, the quality of the final analytical result depends on using the same amount of human DNA in each sample.
- For PCR based reactions, the addition of too much DNA to an amplification reaction is as undesirable as too little.
- The slot blot is the method of choice for determining the amount of human DNA.(Kits: Applied Biosystems Quantiblot, Promega AluQuant)
- Procedure:
- – a small portion of sample is applied to a nylon membrane
- – a set of standard samples , for which quantities are known, is also applied for comparison.
- – samples are permanently fixed to membrane, and probed with small fragment of synthetic DNA selected to hybridise to higher primate DNA.
- – probe is tagged with an enzyme that interacts with a certain chemical to discharge light (Chemiluminescence).
- – probed membrane soaked in chemical and exposed to X-ray film; black band corresponds to region detected by probe.
- Quality
- For RFLP analysis it is important to know how much degraded DNA is in sample and how much total DNA is in sample.
- DNA in large pieces of high molecular weight is critical to obtaining a RFLP result.
- Typically a yield gel is used for determining state of DNA and quantitating total amount of DNA present.
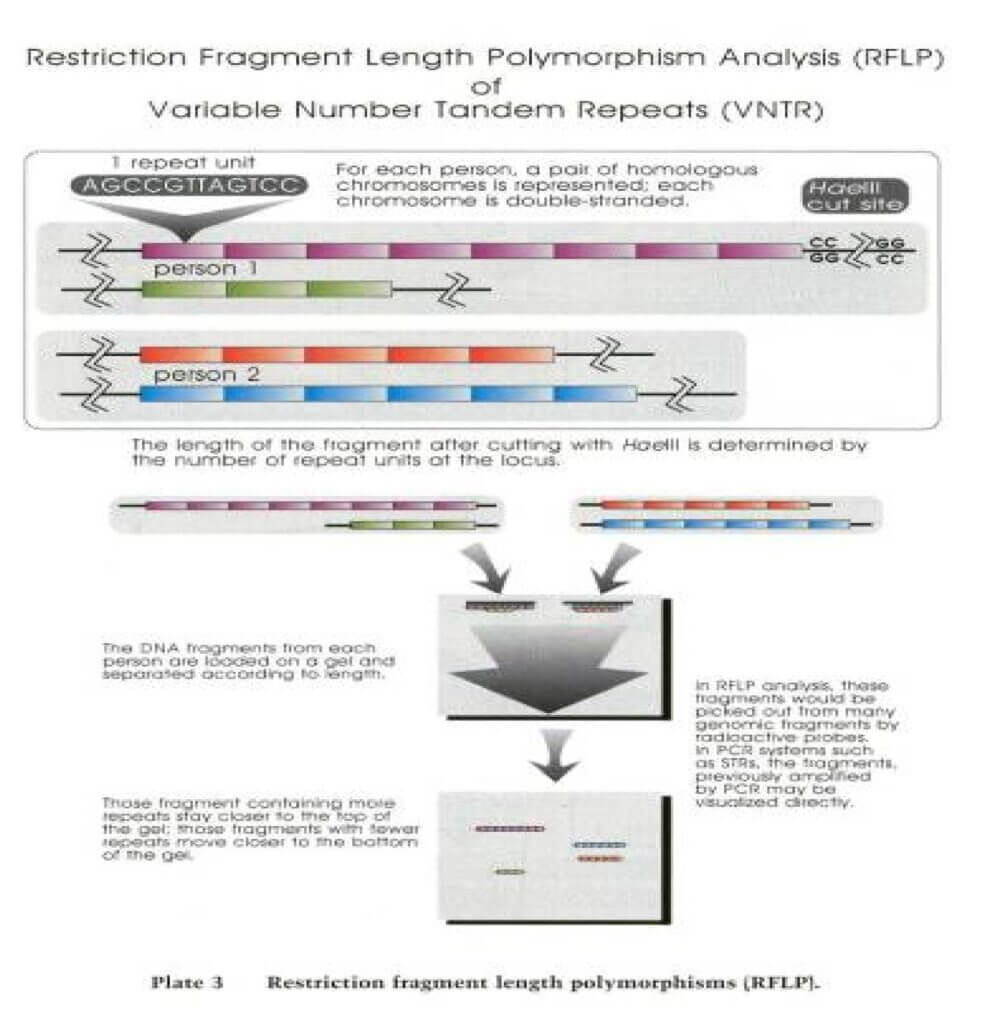
RFLP Analysis
Measures size of DNA fragments produced by Restriction endonucleases.
PCR Amplification
- · Usually performed on samples that are too small or too degraded to give reliable RFLP results.
- · Process faithfully replicates a defined segment of DNA millions of times; it is dependent on Taq polymerase
Analysis of PCR Product
Sequence Polymorphisms
AmpliType PM + DQA1
- Principle of complementary base pairing; under appropriate conditions, only those single strands that match exactly will hybridise. If only a few bases are different, the 2 strands will fail to attach.
- Procedure:
- A nylon strip to which DNA probes have been attached is challenged with PCR product
- These strips are commercially available and contain specific DNA sequences originating from same locus in the genome as the DNA that was amplified by PCR.
- · When probes are attached to the typing strip the format is known as reverse dot blot.
- · Each probe is a specific sequence of DNA that defines an allele
- · These types of probes are known as sequence specific-oligos (SSO), or allele specific-oligos (ASO).
- · The SSO probes on membrane define a finite number of variations seen at this particular region.
- · The type of the sample is revealed by hybridization of the amplified DNA to a specific immobilized probe on the strip.
- a. The primers used in the amplification reaction are labeled with a biological tag called biotin.
- b. After all PCR product has bound to probe strip, excess unbound PCR is removed and streptavidin/HRPO conjugate is applied and binds tightly to biotin. Biotin molecule has strong affinity for streptavidin, and streptavidin is chemically linked to HRPO.
- c. A colourless substrate is added (tetramethyl benzidene, TMB) and HRP releases a blue colour in presence of hydrogen peroxide.
- d. Dots to which DNA has adhered turn blue. The pattern of dots corresponds to alleles present in sample.
- mtDNA
- The variation exhibited in the hypervariable region of the mtDNA chromosomes are generally point mutations.
- Direct DNA sequencing is usually used.
- Sequencing is most basic and comprehensive method of comparing DNA fragments, however not practical on genome samples since most loci selected are highly variable.
- Most people are heterozygous at nDNA loci chosen for forensic analysis; no way to sort out different sequences that should be obtained.
- DNA sequencing is also time consuming, cumbersome, and technically challenging. Disadvantages outweigh advantages and are not method of choice for nDNA.
- mtDNA region is small and easily sequenced
- · only a single locus analysed and only a single allele is present in an individual.
- · most DNA sequence is now performed using one of the automated systems.
- · reactions and analysis steps are the same as manual sequencing eg. Variations of the Sanger method.
- · A different approach is the analysis of mtDNA hypervariable regions involving identification of mutational hot spots and creation of a set of defined probes for those sites
- mtDNA probe strips contain a total of 31 immobilised probes that distinguish variants in 10 segments that span hypervariable region (HVI) and HVII, resulting in 100s of possible types.
- Defined regions of HVI and HVII are amplified and allowed to interact with the strip.
- Where a particular sequence is complementary to the immobilized probe occurs, hybridization takes place and the area turns blue.
- An individual type is read by noting the presence and location of blue lines in each of the strips that correspond to defined alleles in each section of HVI and HVII. Presented in digital format, which is ideal for computerized detection, analysis, storage and comparison.
- Length Polymorphisms (DIS80, STRs, Gender ID)
- Amplifies STRs have now become the system of choice for forensic DNA testing.
- The system for all three detection systems is similar (DIS80, STRs, Gender ID).
- The PCR product is either loaded into a gel or introduced into a capillary containing polyacrlyamide.Polyacrylamide more appropriate for analyzing smaller size PCR products than agarose used in RFLP
- Gel run and PCR fragments are separated by length (plate 8).
- Bands can be stained directly (silver: sidebar 6)
- Gel is dried and kept as a permanent record.
- The pattern of band obtained is compared between samples as well as to an allelic ladder consisting of most or all of the alleles at each locus.
- Each locus will produce 1 or 2 bands representing the alleles present. STRs in particular show a high degree of homozygotes because system is less polymorphic.
- If several loci are amplified in the same tube they are consequently run on the same lane.
- A lane containing the products of three loci may contain from 3 to 6 bands depending on whether the person is homozygous or heterozygous at each locus.
- Each locus has its own allelic ladder for comparison
- Alleles from each locus do not overlap.
- Gender locus often appended to STR multiplex system.
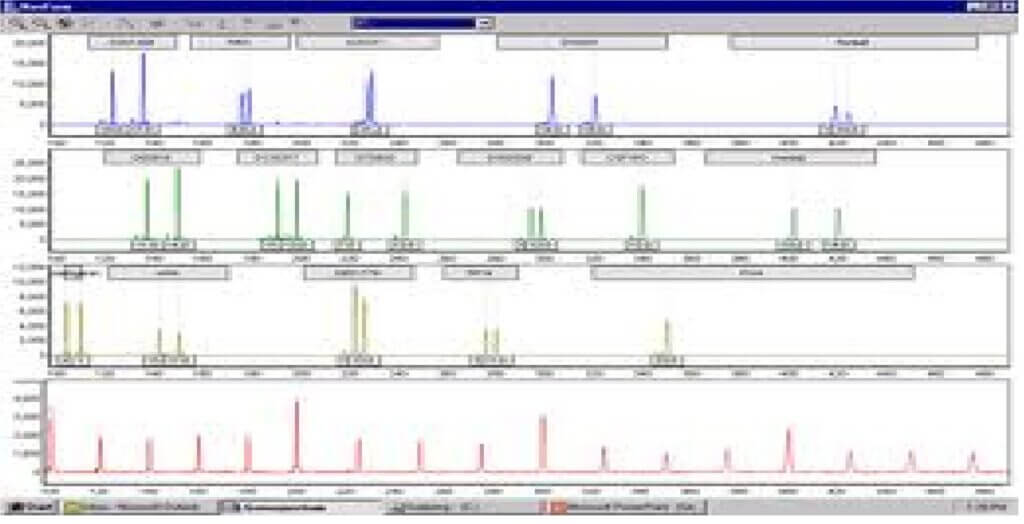
- STRs are now most commonly analysed by fluorescent detection and automated analysis. Two variations to this theme:
- First method: acrylamide gel may be run as for manual analysis but gel is read by computerized laser detection system instead of directly by eye. The results look similar to those obtained using silver staining
- Second method: A laser beam detects bands as they run off the gel or capillary column Results called electropherograms, are usually presented as a series of peaks. All of these automated systems rely on fluorescent tag that is incorporated into PCR product during amplification.
Evidence Collection and Packaging
Please follow the directions below for collecting and packaging evidence for DNA testing.
- Use clean latex gloves for collecting each item of evidence. It is recommended to change gloves between the handling of different items of evidence.
- Each item of evidence must be packaged separately.
- Bloodstains, semen stains, and other types of stains must be thoroughly air-dried and packaged in sealed paper envelopes or paper bags. For proper chain of custody, all packages must be marked with case number, item numbers, and date, and must be initialed across the seals.
- If stains must be transferred from an unmovable surface (such as a window or sidewalk), sterile cotton swabs and distilled water may be used.
- Photograph the surface with a ruler before swabbing. Moisten the swab with water and shake the swab to remove the excess water.
- Rub the stained area with the moist swab until all of the stain is transferred to the swab. If one swab is insufficient to collect all the stain, use additional moist swabs to collect all of the stain.
- Two control swabs may also be collected as controls for other serological tests:
(Swab 1) Swab an unstained area adjacent to the stained area using a moist swab.
(Swab 2) Provide a moist swab with nothing else on it but the water used in the collection process. - Prepare properly marked envelopes (such as coin envelopes) or paper containers for the swabs.
- Air dry the swabs without permitting the swabs to touch one another. If time requires, the swabs may be placed still moist in the envelopes until they can be transported to a place where they can be properly air dried. (This is why paper containers are preferred and glass or plastic containers should be avoided. Paper containers allow moisture to escape to prevent bacterial degradation of the DNA.)
- Place swabs in appropriate separate paper containers, properly marked for identification.
- Scraping dried stains should only be used instead of swabbing if the surface is perfectly smooth and the scraping will result in almost no loss of material. For example, a stain on a smooth vertical surface can be collected (after photographing with a ruler in the picture) by folding a clean sheet of paper in half and taping the top edge of the paper to the surface directly beneath the stain. With a sterile scalpel blade or unused single-edged razor blade, the stain can be scraped into the fold in the paper. Then carefully remove the paper from the surface, remove the tape, fold the paper into a packet, seal with evidence tape and initial properly.
- Evidence which is incapable of drying such as pieces of tissue, organ, bone, liquid urine, or other biological material, should be packaged separately in an air tight container, sealed and marked properly for identification, and immediately frozen and kept stored frozen until shipment. Never use formalin or formaldehyde to preserve any biological evidence because these chemicals degrade DNA.
- Known standard blood samples from deceased individuals should be transferred by syringe into a purple top tube (contains EDTA), properly marked for identification, placed in a paper container, properly marked and sealed with evidence tape for proper chain of custody, and stored refrigerated until shipping. To avoid breakage, do not freeze. Additional packaging will be needed for protection from breakage during shipping.
- Known standard blood samples from living persons should be drawn in purple top tubes, properly marked for identification, placed in a paper container, properly marked and sealed with evidence tape for proper chain of custody, and stored refrigerated until shipping. To avoid breakage, do not freeze. Additional packaging will be needed for protection from breakage during shipping. Dried blood standards are also acceptable.

This is a topic that is near to my heart… Many thanks!
Exactly where are your contact details though?
Respected sir/mam ,
I am sorry for your inconvenience.
You can find our contact information on the top right corner of any page of this website (“contact us” button).
If you wish to register with us you can also find the register button at the home page. It will be appreciated.
Thank you.
I have been surfing on-line more than 3 hours these days,
but I never found any attention-grabbing article like yours.
It’s beautiful worth sufficient for me. Personally, if all webmasters and bloggers made excellent content material as you did, the web shall be much more helpful than ever before.
Thank you very much for your comment and if you want to register it would be appreciated.
If you want to obtain a great deal from this piece of writing
then you have to apply such strategies to your won website.
Feel free to surf to my web-site: delta 8 THC gummies – http://www.thedailyworld.com,
Superb site you have here but I was curious if you knew of
any message boards that cover the same topics discussed in this article?
I’d really like to be a part of group where I
can get feedback from other knowledgeable individuals that share the same interest.
If you have any suggestions, please let me know. Thanks!
Also visit my blog cannabis gummies
Wow that was strange. I just wrote an extremely long
comment but after I clicked submit my comment didn’t show up.
Grrrr… well I’m not writing all that over again. Anyhow, just wanted to say superb blog!
My web page: marijuana gumies
WOW just what I was searching for. Came here by searching for best
THC gummies
my blog weed edibles
I will immediately seize your rss as I can’t to find your e-mail subscription hyperlink or e-newsletter service.
Do you have any? Please let me understand so that I could subscribe.
Thanks.
Feel free to surf to my website; cannabis gummies
Thank you very much for your comment and if you want to register it would be appreciated.
https://informaticss.com/contact-us
https://informaticss.com/users-write-won-pen
https://informaticss.com/register
Email is informaticspvt@gmail.com and divya@informaticss.com
I every time used to read piece of writing in news papers but now as I am a
user of web thus from now I am using net for articles, thanks to web.
Feel free to surf to my blog Observer
Have you ever thought about adding a little bit more than just your articles?
I mean, what you say is fundamental and all. However just imagine if you added some
great pictures or videos to give your posts more, “pop”!
Your content is excellent but with pics and video clips,
this site could undeniably be one of the best in its niche.
Superb blog!
My web blog weed near me
I’m now not sure where you’re getting your info, however great
topic. I must spend some time studying much more or understanding
more. Thanks for wonderful information I was looking for this info
for my mission.
Also visit my site: weed dispensaries (Alphonse)
It’s going to be end of mine day, except before end I am
reading this wonderful post to increase my know-how.
I like the helpful info you supply on your articles. I’ll bookmark your blog and check again right here
frequently. I’m moderately sure I will be informed plenty of new stuff right right here!
Good luck for the following!